Going viral –
Some viruses have genomes larger than some bacteria, can do gene editing.
pm UTC
One of the viruses had a gene that was over 2, bases long — 1.5 times the size of the entire genome of some small viruses. With the assembly complete, the researchers started comparing sequences to figure out what these viruses were related to. In many cases, the answer turned out to be “each other.” The largest viruses were all part of a family that the researchers termed “Mahaphages” (Maha being the Sanskrit word for huge). Significantly, there were no small viruses that grouped among the giants, indicating that these huge genomes are probably stable features of this family rather than being the result of a smaller virus that happened to gain a lot of extra DNA recently.
Many of these viral families have genes for the transfer RNAs used in making proteins, which are normally supplied by the cell. Other genes include those needed for the metabolism of nucleic acids, allowing them to make some of the DNA and RNA they’re dependent on. Normally, both of these classes of genes are provided by the host, although similar things are found in the giant viruses that infect amoeba. The authors note that this sort of gene content is similar to a group of tiny bacteria with small genomes that are thought to be symbiotic or parasitic. Whether this is simply a consequence of lifestyle or represents something more significant is left to future studies. CRISPR-y Many of the viruses also carry components of the CRISPR / Cas system that we’ve started using for genome editing. Bacteria typically use this system to protect themselves from viruses, which makes it odd to find viruses carrying their own version. Some of these systems seem to target genes that bacteria use to control gene activity, so the virus’ version may simply involve redirecting these control systems to focus on virus production. In other cases, they target different viruses, suggesting they’re a way of limiting competitors.
Other families of viruses seem to carry proteins that shut the bacterial CRISPR system down, which is more in line with what you ‘ d expect — a means of protecting the virus from the host’s defenses.
Perhaps the strangest things found in these viruses are genes that encode relatives of a protein called tubulin, which helps a cell organize its internal contents. Bacteria are rather notable for having a poorly defined internal organization, so seeing a virus leveraging something we don’t understand especially well is rather striking. Still, it’s easy to see how this protein could help get all the pieces needed for assembling a virus to the right place. 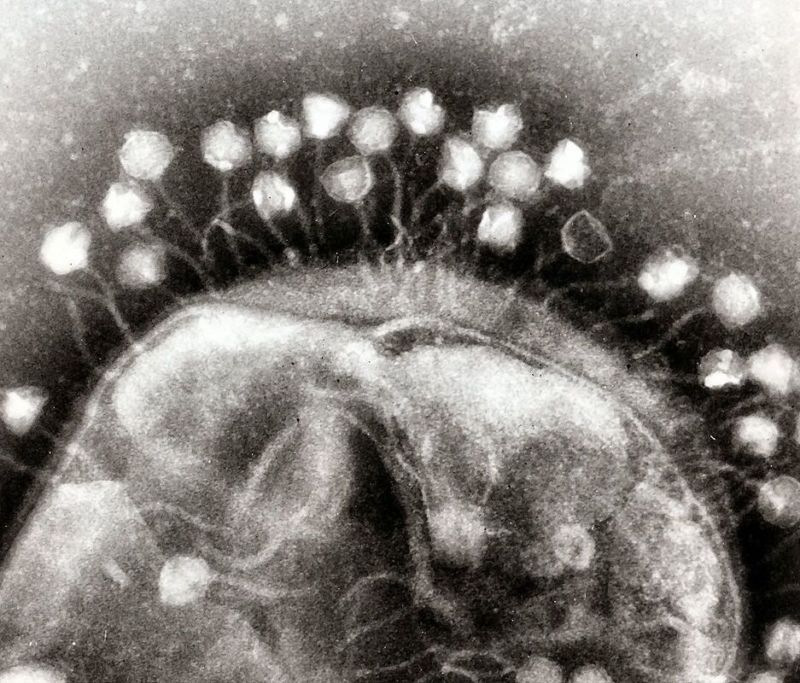 But there’s clearly a lot we don’t understand about these viruses more generally, including the specific cells they infect — we know the environment they came from and the genuses of bacteria they’re generally found with, but not a whole lot more than that. Figuring out more and studying their dynamics in culture may help us understand how they can sometimes outcompete their smaller and faster-moving relatives. And, in the process, might teach us some lessons about the bacteria they’re infecting.
But there’s clearly a lot we don’t understand about these viruses more generally, including the specific cells they infect — we know the environment they came from and the genuses of bacteria they’re generally found with, but not a whole lot more than that. Figuring out more and studying their dynamics in culture may help us understand how they can sometimes outcompete their smaller and faster-moving relatives. And, in the process, might teach us some lessons about the bacteria they’re infecting.




GIPHY App Key not set. Please check settings